Professors Guoqiang Bi and Beiming Liu from the University of Science and Technology of China, Hefei National Research Center for Microscale Material Science and the School of Life Sciences and Medicine, in collaboration with the team from the Hefei Comprehensive National Science Center's Artificial Intelligence Research Institute and the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, have made major breakthroughs in the field of large-scale biological tissue 3D microscopic imaging. The team has successfully developedThe world's fastest subcellular-level high-definition 3D imaging technology for small animals, achieving the efficient mapping of the fine atlas of the peripheral nervous system. Related results were published on July 10 in "Cell" (Cell)on.
3D visualization of peripheral nerve structures in the mouse body
The peripheral nervous system, as the body's "Internet of Things", carries the bidirectional communication and regulation tasks between the brain and all organs: on the one hand, it transmits movement instructions and regulates critical functions such as breathing and heartbeat; on the other hand, it real-time feedbacks sensory signals such as pain and temperature to the central nervous system for processing, thereby coordinating the activities of various tissues and organs. Mapping the fine connectivity of the peripheral nervous system throughout the body is key to understanding its complex functional mechanisms and related disease mechanisms.
For a long time, scientists' understanding of the overall structure of the peripheral nervous system relied on millimeter-resolution anatomical studies. Although advances in three-dimensional optical microscopy over the past decade have promoted the解析 of brain-wide mesoscopic neural maps at the micron level, research on the peripheral nervous system throughout the body still faces technical bottlenecks. Existing advanced imaging technologies find it difficult to simultaneously achieve high resolution and high imaging speed. Even with the transparent processing of whole-body samples, it is still challenging to解析 the complex long-range pathway structures of the peripheral nervous system throughout the mouse body at the subcellular resolution.
The research team previously developed a new synchronous fly-scanning technology (VISoR), which combines thick slicing and clearing of large biological samples for three-dimensional microscopic imaging. VISoRCombines high speed, high resolution, and scalability, enabling sub-micron resolution imaging of mouse whole-brain samples within 1.5 hours (Hao Wang, Qingyuan Zhu, et al., "National Science Review", 2019), and after further optimization, it achieved micron-resolution 3D imaging and single-neuron fiber tracing of the macaque whole brain for the first time (Fang Xu, Yan Shen, Lufeng Ding, Chaoyu Yang, et al., "Nature Biotechnology", 2021). However, this whole-brain imaging strategy, which involves slicing before clearing, is not suitable for whole-body mouse samples. Unlike the relatively dense and homogeneous brain, the whole-body tissues of mice have high heterogeneity, containing diverse tissue types and irregular structures, making them prone to tissue dislocation and loss during the slicing process, thus hindering complete reconstruction.
To address this technical challenge, the team proposedThe strategy of "sample in situ sectioning + sectioned surface 3D imaging" and the development of the blockface-VISoR imaging system integrated with a precision vibration sectioning device, and the supporting sample preparation process of whole-body clearing and hydrogel embedding for mice, ARCHmap. The core of this technical process is to perform 3D imaging only on the surface of the sample block to a depth of about 600 microns each time, then automatically excise the imaged 400-micron-thick sample, and cycle this process until the sample imaging is complete. Then, using an automated stitching algorithm, the overlapping areas of about 200 microns between adjacent slices are seamlessly stitched and reconstructed in 3D. Since the imaging depth for each scan is only a few hundred microns, the light scattering effect after tissue clearing is weak, so high-resolution imaging can be achieved. Based on this strategy, researchers have established an optimized technical process,Completed uniform subcellular resolution 3D imaging of the entire body of adult mice within 40 hours, acquiring about 70 TB of raw image data per channel. So far, data from dozens of mice have been collected, totaling over 4 PB.

blockface-VISoR imaging system
Due to the advantages of the sample preparation method with high fluorescence preservation, the ARCHmap-blockface-VISoR technologyCompatible with commonly used transgenic and neurotropic virus-carried fluorescent proteins in the field of neuroscience, as well as immunofluorescence and other labeling methods. By combining the above labeling and imaging techniques, researchers have successfully resolved the fine structures and single-fiber projection pathways of different types of peripheral nerves throughout the mouse body. This study reveals, for the first time, the segmental projection characteristics of single spinal neurons, clarifies the organ-specific vascular distribution patterns of sympathetic nerves throughout the body, and解析了迷走神经的整体投射构架and单纤维复杂投射路径.
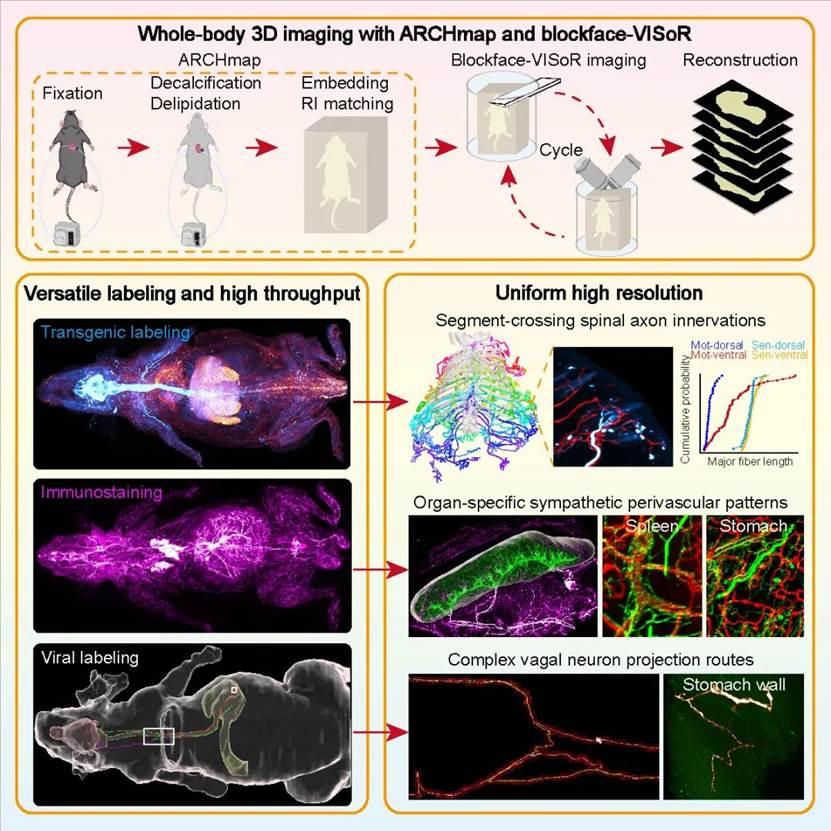
Mouse whole body blockface-VISoR imaging process and surrounding nerve mesoscopic structure analysis
Researchers say this breakthrough technology helps establish new paradigms for studying peripheral nerve connectomics and understanding neural regulation structures. It also has important applications in developmental biology, systemic anatomy, and biomedicine. Moreover, there is still room for improvement and optimization. The next step is to use dual or multi-camera imaging for simultaneous multi-channel image acquisition to improve data collection efficiency and explore its application in imaging larger-scale biological samples.


Discussion between mentors and team members
Reviewers of the journal "Cell" highly praised this work, pointing out that "these analyses produced strikingly detailed data at both the population and single-cell levels. Importantly, new insights emerged from this initial exploration," and commented, "This is an interesting work, beautifully illustrated, and the methodology shows great potential."
Team members conducting experiments
Meiyu Shi, a specially appointed associate researcher at the University of Science and Technology of China, doctoral students Yuchen Yao, Miao Wang, and master's student Qi Yang are the co-first authors of the paper. Professor Guoqiang Bi is the corresponding author, with special researcher Xu Cheng, Professor Beiming Liu, and senior engineer Qingyuan Zhu as co-corresponding authors. The University of Science and Technology of China is the primary completion unit for this work, with the Hefei Comprehensive National Science Center's Artificial Intelligence Research Institute and the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences as the second and third completion units, respectively. This work also received collaborative support from Professor Qu Lei of Anhui University, researcher Xu Fuqiang of the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Professor Zhang Ming of the University of Otago, New Zealand, and Professor Yusuf Ozgur Cakmak. In addition, this research was funded by the National Key Research and Development Program, the National Natural Science Foundation, the Science and Technology Innovation 2030 - "Brain Science and Brain-like Research" major project, the Chinese Academy of Sciences' strategic priority research program, the University of Science and Technology of China Youth Innovation Fund, and the Anhui Provincial Natural Science Foundation.

Research team photo
(From left to right: Zhang Keming, Xu Cheng, Qingyuan Zhu, Guoqiang Bi, Beiming Liu, Shi Meiyu, Yuchen Yao, Miao Wang)
Paper Link:
https://www.cell.com/cell/fulltext/S0092-8674(25)00673-7
Links to experimental techniques and demonstration datasets:
https://mesoanatomy.org/mesomouse
Source: USTC News Network
Photography: Xinyu Zhou
Cover: Xingjiaozhe Lv
Editor: Siming Zeng